Hampton Research蛋白结晶试剂盒






Products > Optimization Screens > Proti-Ace > Proti-Ace • Proti-Ace 2
Proti-Ace • Proti-Ace 2
Applications
- Limited proteolysis, in situ proteolysis, and proteolytic screening of protein samples for crystallization and structure determination
Features
- Proti-Ace proteases
- alpha-Chymotrypsin
- Trypsin
- Elastase
- Papain
- Subtilisin
- Endoproteinase Glu-C
- Proti-Ace 2 proteases
- Proteinase K
- Clostripain (Endoproteinase-Arg-C)
- Pepsin
- Thermolysin
- Bromelain
- Actinase E
- Optimized, stable, freeze dried protease formulation
Description
A proteolytic fragment or domain of a protein may crystallize more readily or form better diffracting crystals than the intact protein.1-8
Proteases can be used to generate small, active fragments or domains of the target protein for crystallization.9 The fragment or domain can be used directly for crystallization experiments. Or the proteolytic sample analyzed by gel electrophoresis and/or mass spectrometry for mass and sequence for subsequent cloning, expression, purification and crystallization. Using proteolysis to enhance sample crystallization, the current overall success rate for yielding a deposited crystal structure is currently better than 12%.3
Proti-Ace (HR2-429) contains three aliquots of 6 unique proteases (a-Chymotrypsin, Trypsin, Elastase, Papain, Subtilisin and Endoproteinase Glu-C) and six aliquots of Proti-Ace Dilution Buffer. Each protease is supplied in a stable, lyophilized format in an optimized digest buffer. Simply add water when ready to use.
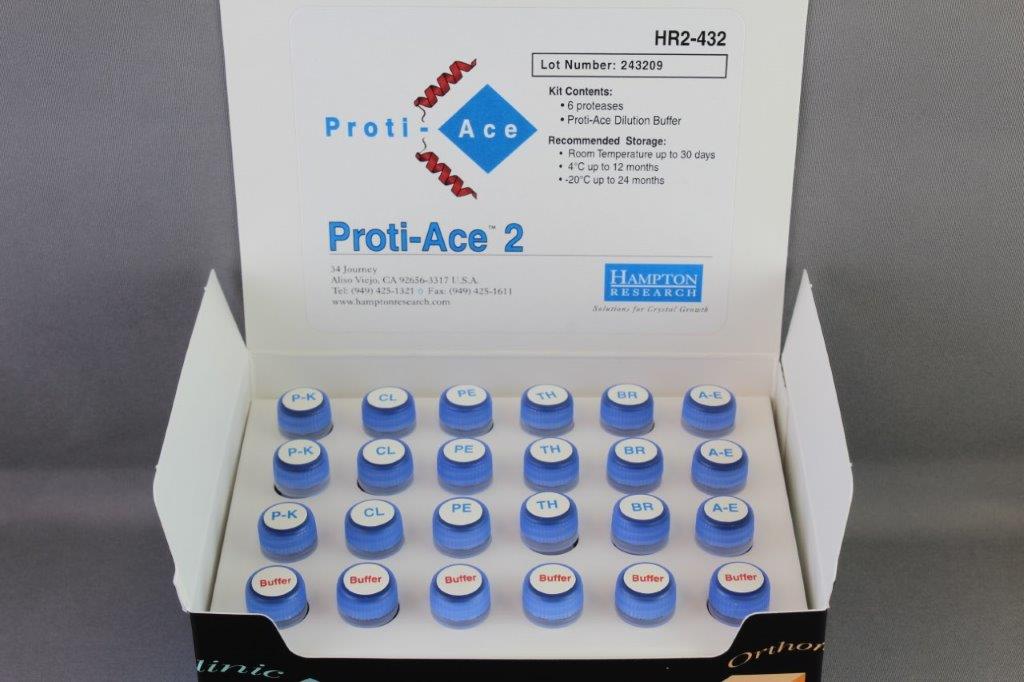
Proti-Ace 2 (HR2-432) contains three aliquots of 6 unique proteases (Proteinase K, Clostripain (Endoproteinase-Arg-C), Pepsin, Thermolysin, Bromelain and Actinase E) and six aliquots of Proti-Ace Dilution Buffer. Each protease is supplied in a stable, lyophilized format in an optimized digest buffer. Simply add water when ready to use.
The unique freeze dried formulation of the Proti-Ace kits offers a much improved protease stability compared to liquid protease formulations.

Click to Zoom In
CAT NO
HR2-429
NAME
DESCRIPTION
tube format
PRICE
$251.00
cart quote
CAT NO
HR2-432
NAME
DESCRIPTION
tube format
PRICE
$251.00
cart quote
Support Material(s)
 HR2-429 Proti-Ace User Guide
HR2-429 Proti-Ace User Guide HR2-429 Proti-Ace SDS
HR2-429 Proti-Ace SDS HR2-432 Proti-Ace 2 User Guide
HR2-432 Proti-Ace 2 User Guide HR2-432 Proti-Ace 2 SDS
HR2-432 Proti-Ace 2 SDS Proti-Ace Enzyme Mr pI Specificty Source
Proti-Ace Enzyme Mr pI Specificty Source Related Item(S)
- Individual Proti-Ace & Proti-Ace 2 Reagents
References
1. Allan D′Arcy, personal communication, 1989-2009.
2. In situ proteolysis for protein crystallization and structure determination. Dong, A et al. Nature Methods – 4, 1019 – 1021 (2007).
3. In Situ Proteolysis to Generate Crystals for Structure Determination: An Update. Amy Wernimont, Aled Edwards. PLoS ONE 4(4): e5094. doi:10.1371/ journal.pone.0005094.
4. The use of in situ proteolysis in the crystallization of murine CstF-77. Tong et al. Acta Cryst. (2007). F63, 135-138.
5. A brief history of protein crystal growth. McPherson, A. Journal of Crystal Growth, vol. 110, issue 1-2, pp. 1-10, 1991.
6. Preparation and analysis of protein crystals. McPherson, A. John Wiley & Sons, Inc. 1982. ISBN 089464355X.
7. A crystallizable form of the Streptococcus gordonii surface antigen SspB C-domain obtained by limited proteolysis. Forsgren et al. Acta Cryst. (2009). F65, 712-714.
8. Preliminary X-ray analysis of a human VH fragment at 1.8 angstrom resolution. Gaur, Kupper, Fischer & Hoffman. Acta Cryst. (2004). D60, 965-967.
9. Replication Protein A Characterization and Crystallization of the DNA Binding Domain. Pfuetzner et al. The Journal of Biological Chemistry, Vol. 272, No. 1, Issue of January 3, pp. 430-434, 1997.
10. Combining in situ proteolysis and mass spectrometry to crystallize Escherichia coli PgaB. Little et al. Acta Cryst. (2012) F68, 842-845.
11. Proteolysis of Native Proteins – Trapping of a Reaction Intermediate. Chenyi Wu, Duncan H. L. Robertson, Simon J. Hubbard, Simon J. Gaskell and Robert J. Beynon. doi: 10.1074/jbc.274.2.1108 January 8, 1999 The Journal of Biological Chemistry, 274, 1108-1115.
12. Crystallization of mouse RIG-I ATPase domain: in situ proteolysis. Civril F, Hopfner KP. Methods Mol Biol. 2014;1169:27-35. doi: 10.1007/978-1-4939-0882-0_3.
13. Crystallization and preliminary X-ray crystallographic analysis of YfcM: an important factor for EF-P hydroxylation. K. Kobayashi, T. Suzuki, N. Dohmae, R. Ishitani and O. Nureki. Acta Cryst. (2014). F70, 1236-1239 [ doi:10.1107/S2053230X14015726 ]. Synopsis: in situ proteolysis crystallization method produces crystals diffracting to 1.45 Å


Hampton Research, first in crystallization since 1991, developing and delivering crystallization and optimization screens, reagents, plates, and other tools for the crystallization of biological macromolecules, including proteins (antibody), peptides (insulin), and nucleic acids (DNA).
- Products
- Gallery
- My Account
|
|
|
- Contact Us
- Quick Order
- Support
|
- Privacy Policy
- Terms and Conditions
|
- Products
- Gallery
- My Account
- Support
- Contact Us
- Quick Order
- Privacy Policy
- Terms and Conditions
|
|
|
|
|
|
|
© 2021 HAMPTON RESEARCH CORP.
| Website by Skyhound Internet