Hampton Research蛋白结晶试剂盒






Products > Optimize Reagents > Optimize – Salts > Tacsimate pH 4, 5, 6, 7, 8, & 9
Tacsimate pH 4, 5, 6, 7, 8, & 9
Applications
- Crystallization grade Tacsimate for formulating screens or for optimization
Features
- Sterile filtered solution
- Formulated in Type 1+ ultrapure water: 18.2 megaohm-cm resistivity at 25°C, < 5 ppb Total Organic Carbon, bacteria free (<1 Bacteria (CFU/ml)), pyrogen free (<0.03 Endotoxin (EU/ml)), RNase-free (< 0.01 ng/mL) and DNase-free (< 4 pg/µL)
Description
To screen pH as a crystallization variable using Tacsimate one can mix together different ratios of different pH Tacsimate solutions. Refer to the pdf “Mixing Tacsimate Stocks to Screen pH” on this page. Or, one can add a buffer to the crystallizaiton reagent containing Tacsimate. Recommended final buffer concentration in the reagent well is 0.05 to 0.1 M.
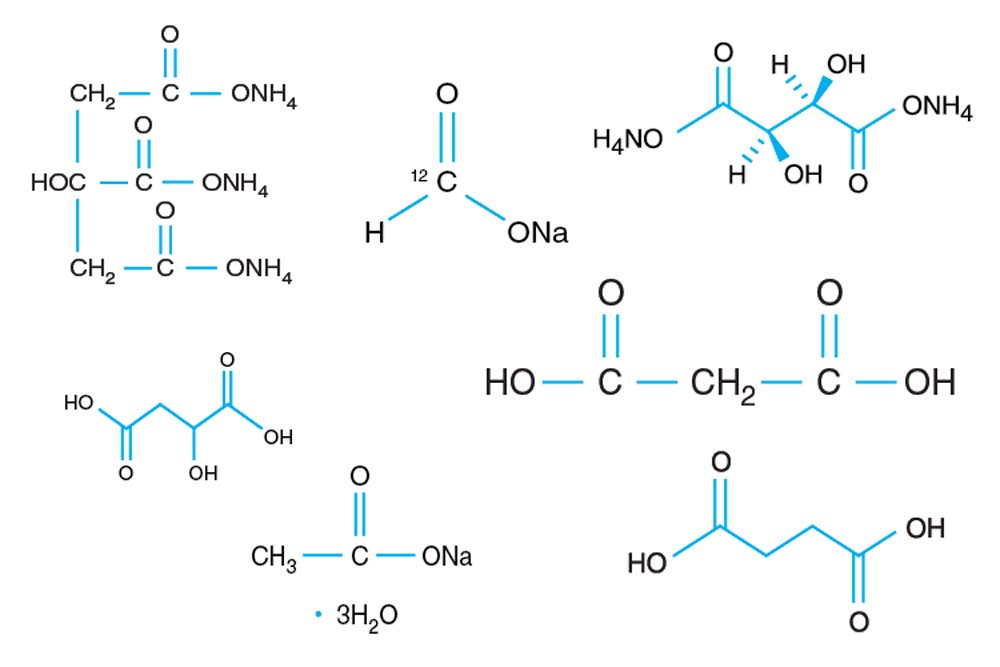
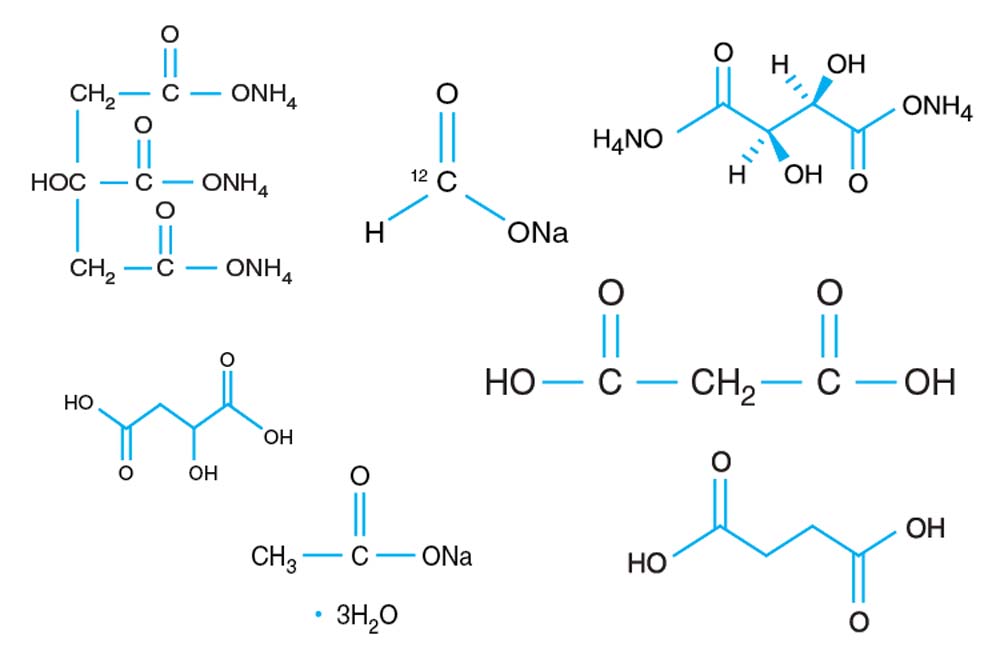
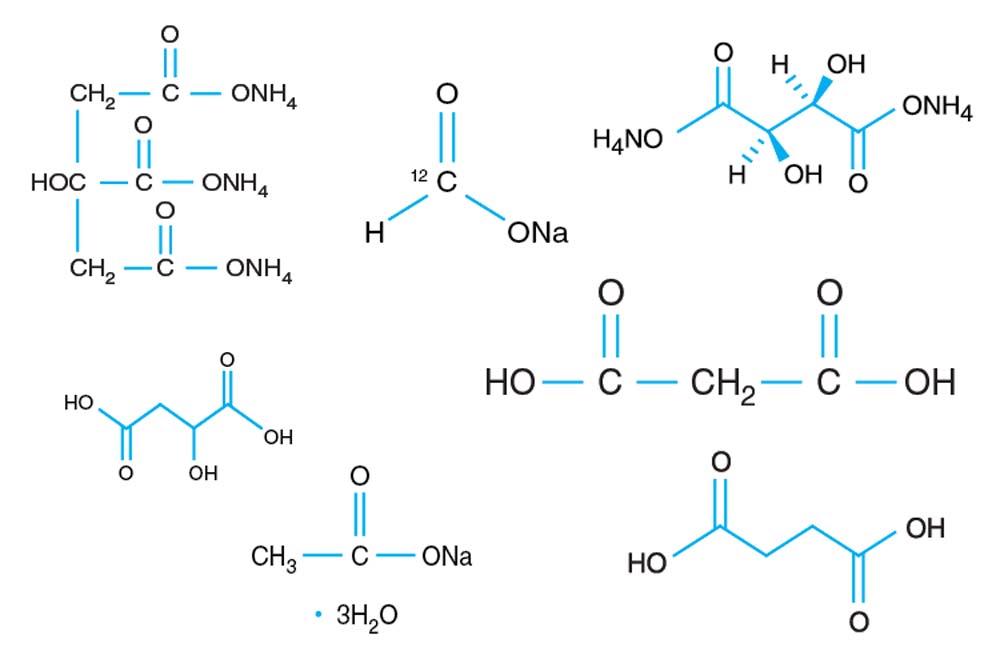
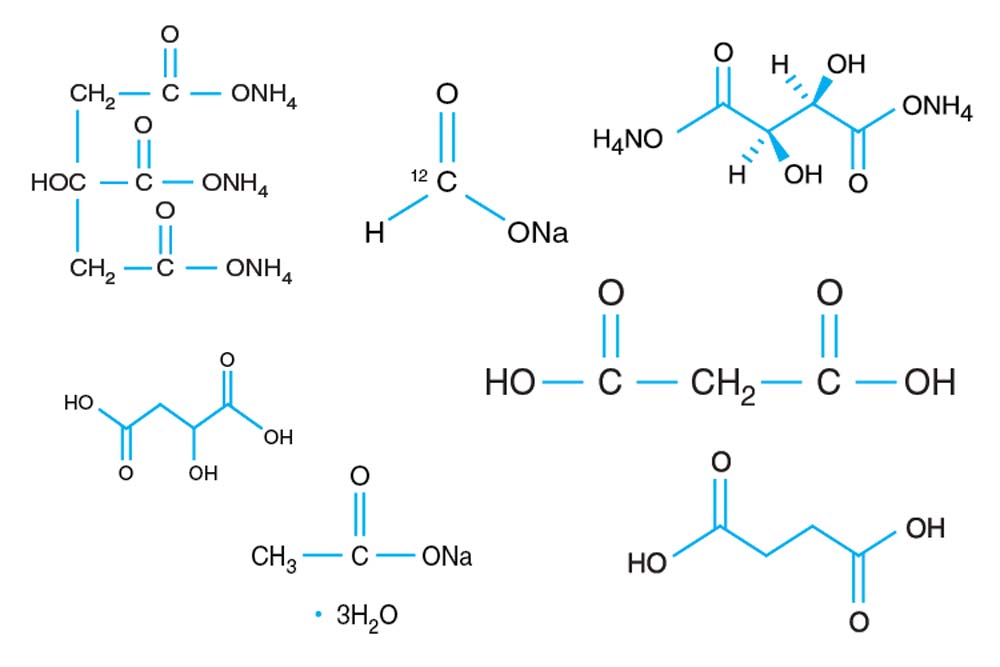
Tacsimate
A pH titrated mixture of organic acids. 1.8305 M Malonic acid, 0.25 M Ammonium citrate tribasic, 0.12 M Succinic acid, 0.3 M DL-Malic acid, 0.4 M Sodium acetate trihydrate, 0.5 M Sodium formate, 0.16 M Ammonium tartrate dibasic. pH adjusted using NaOH
Measured pH of 100% Tacsimate is 4.0 at 25°C
Measured pH of 100% Tacsimate is 5.0 at 25°C
Measured pH of 100% Tacsimate is 6.0 at 25°C
Measured pH of 100% Tacsimate is 7.0 at 25°C
Measured pH of 100% Tacsimate is 8.0 at 25°C
Measured pH of 100% Tacsimate is 9.0 at 25°C
Click to Zoom In
CAT NO
HR2-823
NAME
DESCRIPTION
200 mL
PRICE
$110.00
cart quote
CAT NO
HR2-825
NAME
DESCRIPTION
200 mL
PRICE
$110.00
cart quote
CAT NO
HR2-827
NAME
DESCRIPTION
200 mL
PRICE
$110.00
cart quote
CAT NO
HR2-755
NAME
DESCRIPTION
200 mL
PRICE
$110.00
cart quote
CAT NO
HR2-829
NAME
DESCRIPTION
200 mL
PRICE
$110.00
cart quote
CAT NO
HR2-813
NAME
DESCRIPTION
200 mL
PRICE
$110.00
cart quote
Support Material(s)
 What is Tacsimate?
What is Tacsimate? Mixing Tacsimate Stocks to Screen Ph
Mixing Tacsimate Stocks to Screen Ph HR2-823 Tacsimate 100% solution pH 4.0 SDS
HR2-823 Tacsimate 100% solution pH 4.0 SDS HR2-825 Tacsimate 100% solution pH 5.0 SDS
HR2-825 Tacsimate 100% solution pH 5.0 SDS HR2-827 Tacsimate 100% solution pH 6.0 SDS
HR2-827 Tacsimate 100% solution pH 6.0 SDS HR2-755 Tacsimate 100% solution pH 7.0 SDS
HR2-755 Tacsimate 100% solution pH 7.0 SDS HR2-829 Tacsimate 100% solution pH 8.0 SDS
HR2-829 Tacsimate 100% solution pH 8.0 SDS HR2-813 Tacsimate 100% solution pH 9.0 SDS
HR2-813 Tacsimate 100% solution pH 9.0 SDS Certificate Of Analysis
References
1. Searching for silver bullets: An alternative strategy for crystallizing macromolecules. Alexander McPherson and Bob Cudney. Journal of Structural Biology 156 (2006) 387–406.
2. A novel strategy for the crystallization of proteins: X-ray diffraction validation. Steven B. Larson, John S. Day, Robert Cudney and Alexander McPherson. Acta Cryst. (2007). D63, 310–318.
3. Structure of the catalytic and ubiquitin-associated domains of the protein kinase MARK/Par-1. Eckhard Mandelkow et al. Structure 14, 173-183, February 2006.
4. A comparison of salts for the crystallization of macromolecules. Alexander McPherson. Protein Science (2001), 10:418-422.
5. Crystallization of a newly discovered histidine acid phosphatase from Francisella tularensis. Richard L. Felts, Thomas J. Reilly, Michael J. Calcutt, and John J. Tanner. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006 January 1; 62(Pt 1): 32–35.
6. Structure of the GTPase and GDI domains of FeoB, the ferrous iron transporter of Legionella pneumophila. Petermann et al, FEBS Letters Volume 584, Issue 4, Pages 733-738 (19 February 2010).


Hampton Research, first in crystallization since 1991, developing and delivering crystallization and optimization screens, reagents, plates, and other tools for the crystallization of biological macromolecules, including proteins (antibody), peptides (insulin), and nucleic acids (DNA).
- Products
- Gallery
- My Account
|
|
|
- Contact Us
- Quick Order
- Support
|
- Privacy Policy
- Terms and Conditions
|
- Products
- Gallery
- My Account
- Support
- Contact Us
- Quick Order
- Privacy Policy
- Terms and Conditions
|
|
|
|
|
|
|
© 2021 HAMPTON RESEARCH CORP.
| Website by Skyhound Internet